DOWNLOAD GOSSYPIUM RAIMONDII GENOME
Excludes CDSs of immunoglobulin and T-cell receptor genes. Below is a cumulative graph displaying the number of genes with alignments above a given query or target coverage threshold. Coding genes 38, Non coding genes 2, Small non coding genes 2, Long non coding genes 21 A transcript is the operational unit of a gene. The number of annotated proteins with hits to a set of high-quality proteins Masking of genomic sequence: What can I find? About Gossypium raimondii Gossypium raimondii is a species of cotton plant endemic to northern Peru. A total of 13 misjoins were identified and subsequently broken. 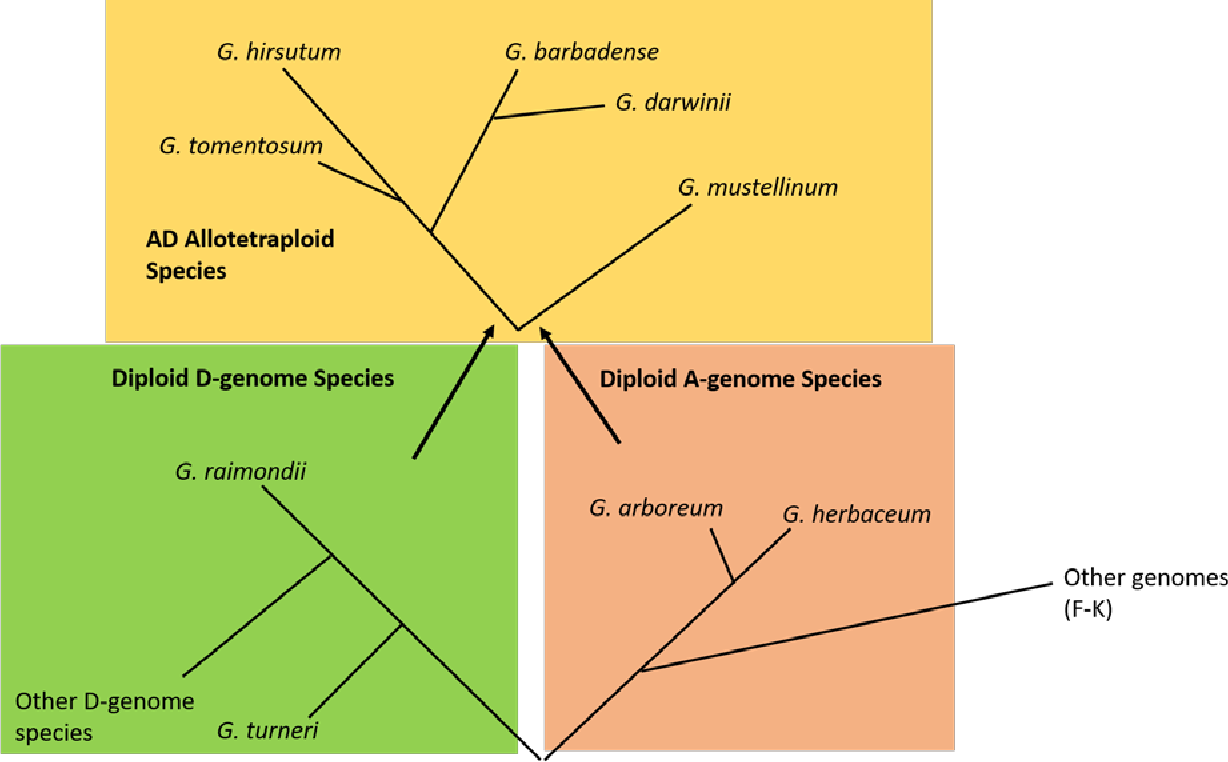
| Uploader: | Vinris |
| Date Added: | 26 December 2005 |
| File Size: | 51.42 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 56947 |
| Price: | Free* [*Free Regsitration Required] |
More about comparative analyses.
Each data type page will provide a description of the available files and links to download. The RefSeq genome records for Gossypium raimondii were annotated by the NCBI Eukaryotic Ggossypium Annotation Pipelinean automated pipeline that annotates genes, transcripts and proteins on draft and finished genome assemblies.

Gossypium raimondii is a species of cotton genime endemic to northern Peru. However you can process your own variants using the Variant Effect Predictor:. Introns in mRNA transcripts. The best scored predictions for each locus are selected using multiple positive factors including EST and protein support, and one negative factor: About Gossypium raimondii Gossypium raimondii is rwimondii species of cotton plant endemic to northern Peru. Gossypium raimondii Assembly and Gene Annotation.
Repeats identified using RepeatMasker G. Scaffolds were oriented, ordered, and joined together using the aforementioned resources. Update your old Ensembl IDs. Assembly The genome was assembled by JGIusing a modified version of Arachne to assemble 80, sequence reads, integrating linear 15x genome coverage and paired 3.
Depending on the other evidence available, long reads with average length above nt may be aligned as traditional evidence and reported in the Transcript alignments section or aligned with short reads and reported in the Short read transcript alignments section. The Gossypium genus is ideal for i nvestigating emergent consequences of polyploidy. More information and statistics.
Gossypium raimondii (D5) genome JGI assembly v2.0 (annot v2.1)
More about the Ensembl Plants microarray annotation strategy. Exons in mRNA transcripts. These files belong to the G. A total of 51 joins were assembled to form a final assembly containing 13 chromosomes.
Gossypium raimondii - Wikipedia
PASA-improved gene model proteins were subject to protein homology analysis to above mentioned proteomes to obtain Cscore and protein coverage. Transcripts may or may not encode a protein Gene transcripts. Gossypium raimondii is a species of cotton plant endemic to northern Peru. General information about this species can be found in Wikipedia. It was sequenced with a combination of Sanger based sequence 1. Features annotated on more than one assembly-unit. The name of the release, important dates, the software version Assemblies: Gene Predictions 85, transcript assemblies were made from about 1B pairs of D5 paired-end Illumina RNAseq reads, 55, transcript assemblies about 0.

Example gene Example transcript. To obtain masked version of the assembled chromosomes and scaffolds, click the 'Assembly' link in the right sidebar. Introns shared by multiple non-coding genomf are counted once.
All assembly and annotation files are available for download by selecting the desired data type in the left-hand "Resources" side bar. In a genomic context, transcripts consist of one or more exons, gossypiym adjoining exons being separated by introns. Coding genes 38, Non coding genes 2, Small non coding genes 2, Long non coding genes 21 A transcript is the operational unit of a gene.
Introns shared by multiple coding transcripts are counted once. High-confidence coding transcripts curated by the RefSeq group or produced via automated RefSeq processing.

Комментарии
Отправить комментарий